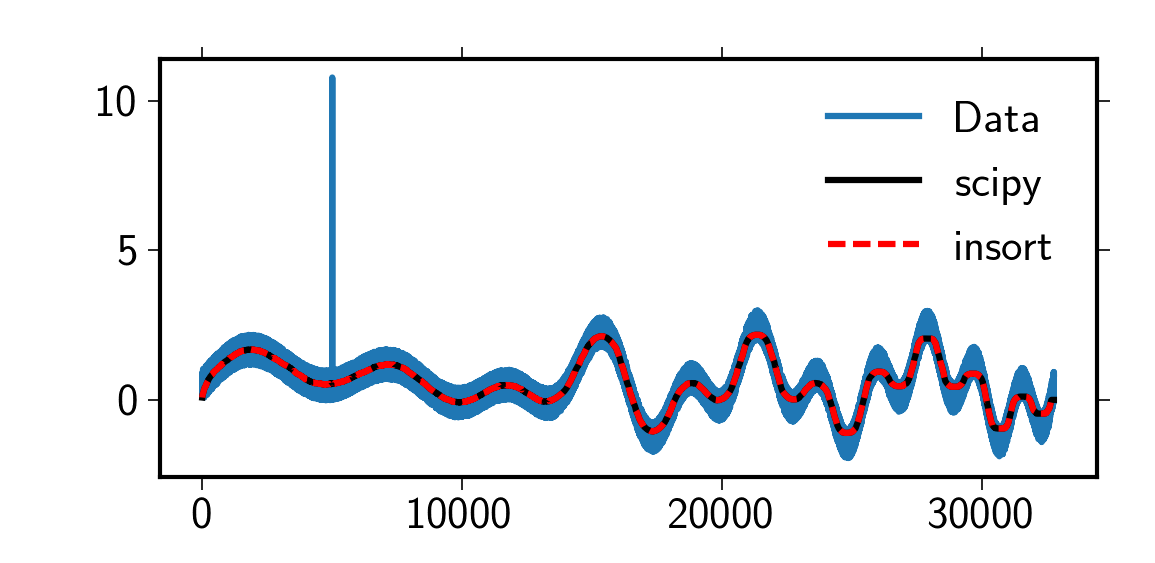
Performance of scipy.signal.medfilter
The median filter in scipy.signal is not optimal. Following
this
discussion, there are much faster implementations, even in pure python. However, the fastest method
used here, running_median_insort, is not a drop-in replacement for
scipy.signal.medfilt as it does not calculate the central median but the
right sided median. However, by zero-padding as in the code below the behavior of the
scipy implementation is exactly reproduced.
def running_median_insort(seq, window_size):
"""
Based on https://bitbucket.org/janto/snippets/src/tip/running_median.py
"""
result = zeros_like(seq)
padded_seq = zeros(len(seq) + window_size)
padded_seq[window_size//2+1:window_size//2+1+len(seq)] = seq
d = deque()
d.extend(padded_seq[:window_size])
s = padded_seq[:window_size]
s.sort()
s = s.tolist()
seq = iter(padded_seq[window_size:])
m = window_size // 2
c = 0
for item in seq:
old = d.popleft()
d.append(item)
del s[bisect_left(s, old)]
insort(s, item)
result[c] = s[m]
c+=1
return resultApplying the filter with width 1023 on an example of some 2^15+1 data samples takes 50.3 ms on my laptop, compared to 2.65s for the medfilter in scipy v0.18 while there is no difference between the results.